Tape Station Results Samples 401, 759, 419
Tape Station Reports - Locus Estimation (500ng DNA)
Ideal number of fragments: 20,000-30,000 Ideal bp range: 75-100, Max bp is 600
Locus Estimation
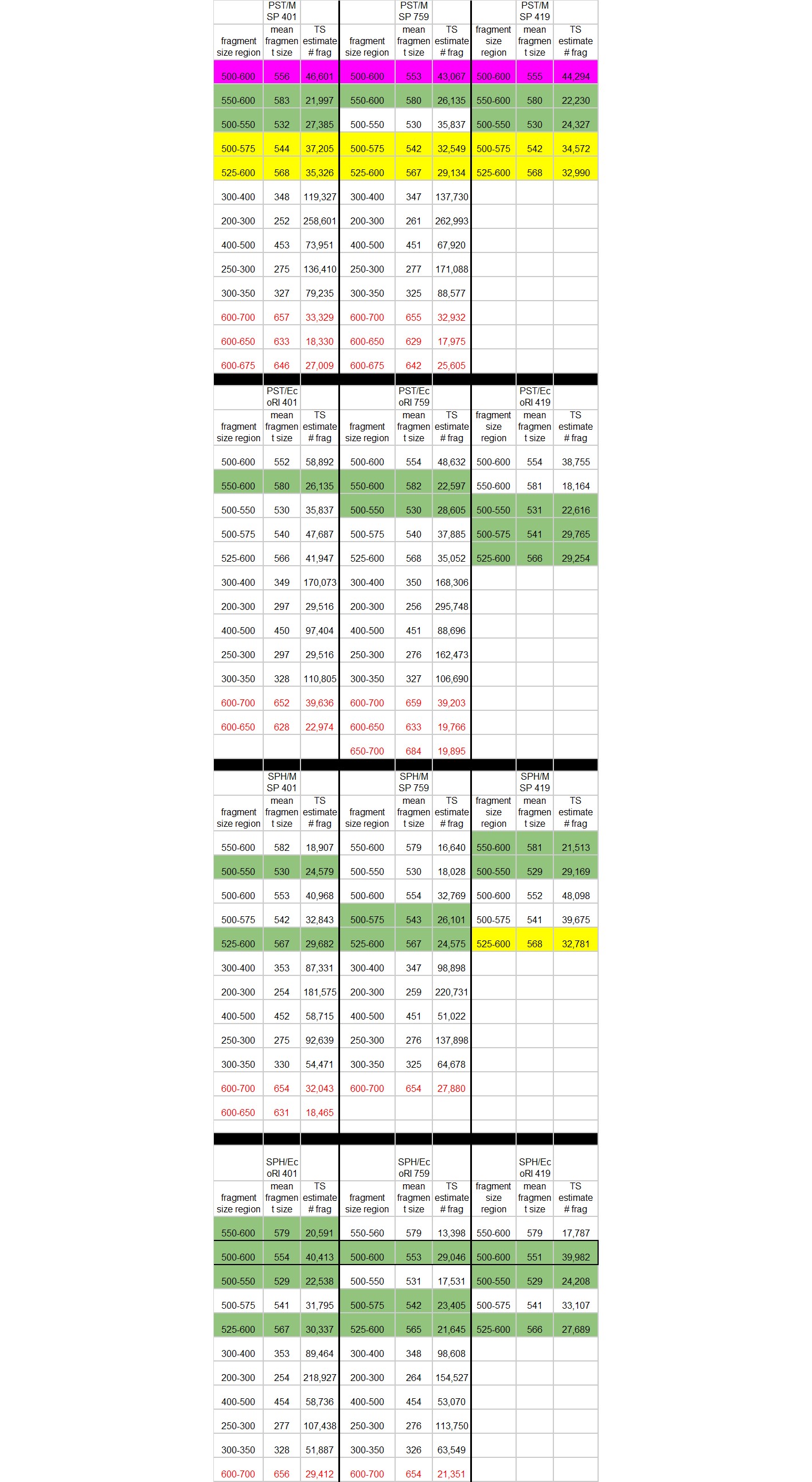
Methods for locus estimation table: Locus Count Estimation (on TS software)
- It’s easiest to do this on the TapeStation computer, you have to use the software that’s on there to look at the files from the machine
- Create a spreadsheet like this (https://docs.google.com/spreadsheets/d/1ojh6jsa2-sxsmnAhJaktRETk4f6ND2gX8wUXyJ2WuS8/edit#gid=0) in your project folder and call it “Locus Count Estimation” Copy columns A-E, rows 1-15 to your spreadsheet, this brings along the calculations with it
- You’ll be making one of these in separate tabs for each enzyme pair for each test sample, a total of 12 (4*3). Label each tab with the sample name and enzyme pair
- Start with just one sample and 1 enzyme pair
- Add in the genome size estimate into field B3
- Open up the files from the TapeStation runs, go to File Explorer -> Documents/Agilent/TapeStation Data/ then find the date you did the TapeStation and the files should be in that folder
- These open up in the TapeStation analysis software (don’t use the “report” PDF)
- Find the wells in the TS that have the sample you’re starting with
- Set fields B8 and B9, this is done once for each enzyme pair and sample (ie once per tab in the sheet)
- Choose 1 enzyme of the pair and go to the single digest well using the TapeStation Region viewing window
- Click on Region Settings. Edit the region to go from 150-60000bp. Check “this file” and click apply
- In the table below, use “Average Size [bp]” to add into your spreadsheet in field B8
- Repeat region for the second enzyme in the double digestions (same sample) and put that average size in the B9 field in the spreadsheet
- You can make note on fields I8 and I9 which enzyme is which if you want to
- Make a table underneath the calculation part in the spreadsheet that has column names “fragment region” “mean fragment size” and “estimated fragments”
- Go to the region viewing window for the double digest of those two enzymes for your sample
- Go to Region Settings and edit the region to be a range between 150 and 600bp, that is not less than 75bp, and not more than 400bp
- Make sure “this file” is checked and click apply
- On the table below the figures, look at average size. Put that number in field B4 in the spreadsheet
- On the table on the software, also look at “% of total”, put that number into a decimal percent (ex. if it says 5.6 you want to use 0.056) in field B2 in the spreadsheet
- On your mini table underneath the calculations, put in your fragment size range and the mean fragment size
- For estimated fragments put in the calculation from field B14 (after all the above numbers are put into the calculation)
- Repeat these steps for as many fragment regions as you want for these enzymes/samples, then move on to do the same for the other enzyme pairs and the other samples
- Remember you are aiming for something that comes out around 20,000-30,000 fragments in the estimated fragments section
Written on February 25, 2021