Summary of all Plate Reader, Qubit, and Tape Station Tests
Summary of Tests
The purpose of this post is to summarize all of the tests I ran to figure out why I was getting different reads (in ng/ul) from the Qubit and Plate Reader. Some of this is duplicate information from other posts, but I think it is important to have it all in one place.
Here is the original protocol for the Biotium Broad Range AccueBlue dsDNA Quantification Assay: https://biotium.com/wp-content/uploads/2017/11/PI-31007.pdf This protocol was edited ONLY to use 2ul of sample/well instead of 10ul of sample/well
Initial Qubit reads for Row A of Plate B post 1.5X bead PRE digest
| Sample | Run 1 |
|---|---|
| S1 | 191.21 |
| S2 | 22154.07 |
| 1 | 6.48, 6.76 |
| 2 | 6.60, 7.07 |
| 3 | 5.70, 5.90 |
| 4 | 5.88, 6.16 |
| 5 | 4.50, 4.70 |
| 6 | 2.52, 2.62 |
| 7 | 7.02, 5.76 |
| 8 | 5.62, 6.24 |
Qubit reads post digest and another 1.5X clean (higher values because more concentrated due to clean)
Post 1.5X Clean Qubit Reads
| Sample | Run 1 |
|---|---|
| S1 | 181.22 |
| S2 | 22197.32 |
| 1 | 9.20 |
| 2 | 8.90 |
| 3 | 7.34 |
| 4 | 7.92 |
| 5 | 6.08 |
| 6 | 3.40 |
| 7 | 7.26 |
| 8 | 7.22 |
Plate reader results following original protocol with 10ul standard/well and 2ul sample/well
| Sample | ng/ul |
|---|---|
| 1 | 26.7 |
| 2 | 21.6 |
| 3 | 12.8 |
| 4 | 17.8 |
| 5 | 14.0 |
| 6 | 6.4 |
| 7 | 14.0 |
| 8 | 11.5 |
Clearly the plate reader values are much higher than the Qubit values. On average, the plate reader predicted a 1.9X higher concentration.
The next thing I tried was using a different species (Crassostrea virginica), the results were similar.
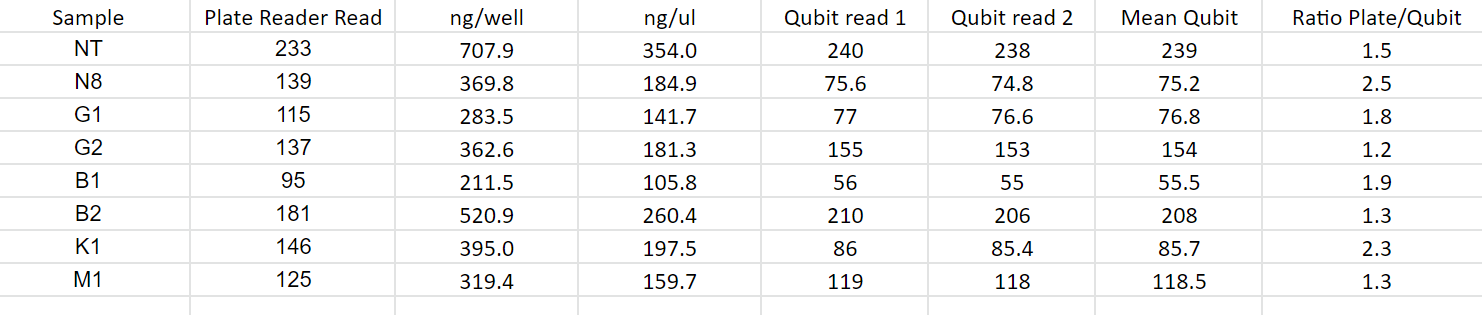
Next I tried two more tests. One where I used a 1/5 ratio so I would be using only 2ul of standard and 2ul of sample so each well would have the same volume. The other test added 8ul of TE buffer to each sample well so all well volumes were the same (210ul). I also ran the samples on the tape station for comparison as this should be the most accurate test. Results below:
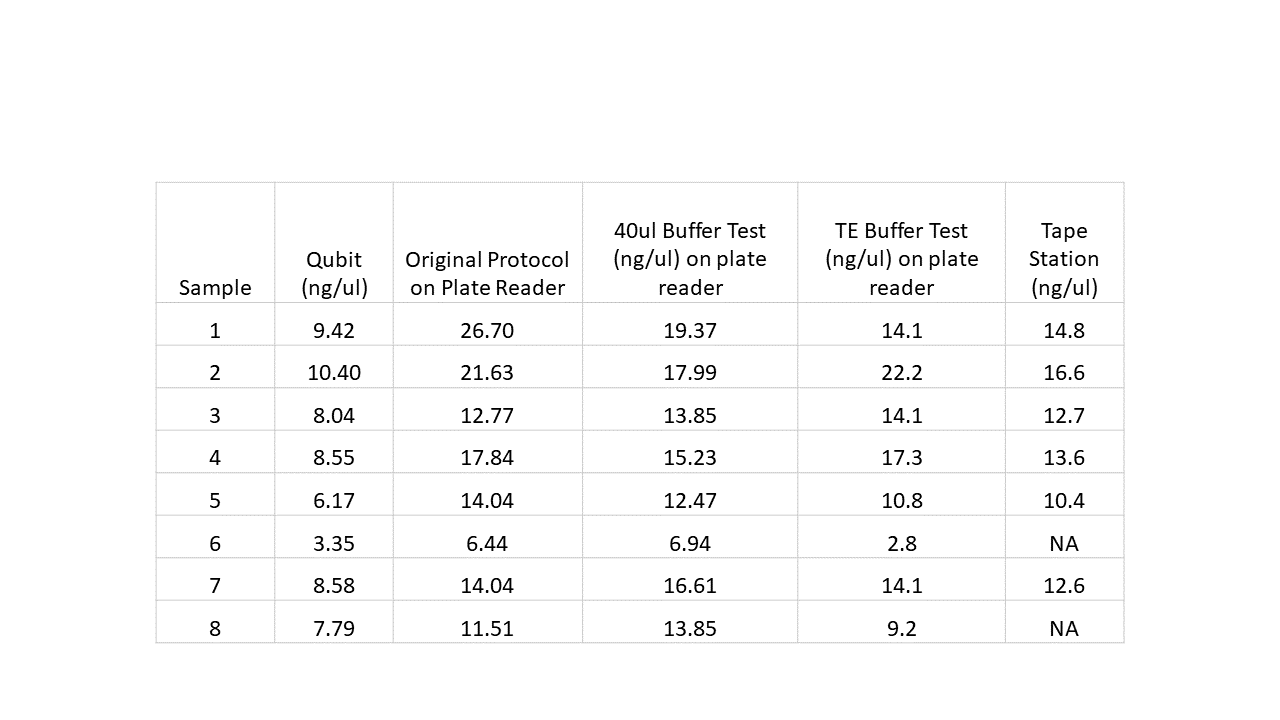
Given these results, I am moving forward with the TE buffer method. I should also note I contacted the manufacturer of the plate reader we have and this is the solution they recommended as well.