Post Digestion Bead Clean and Plate Read
Plate C Post Digest Clean and Plate Read
Date Performed: April 27th, 2021
Followed Step 7 From DDRad protocol for post digestion 1.8X bead clean on plate C
https://docs.google.com/document/d/1iWBGgBZuXlVuiEV-A3ecxeyn9ZaY389tAqGZWksjs5Y/edit
Followed Protocol for Plate Reader:
https://docs.google.com/document/d/1iWBGgBZuXlVuiEV-A3ecxeyn9ZaY389tAqGZWksjs5Y/edit
For Plate C made the following master mix:\
- 37 samples plus 12 standards
- 10mL buffer
- 100ul dye
- 100ul enhancer
(200ul buffer per sample + 2ul dye per sample + 2ul enhancer per sample plus error, also include each standard you’re going to do here as a sample)
Standard Setup
- 2 0ng/ul
- 2 2ng/ul
- 2 12ng/ul
- 2 25ng/ul
- 1 50ng/ul
- 1 100ng/ul
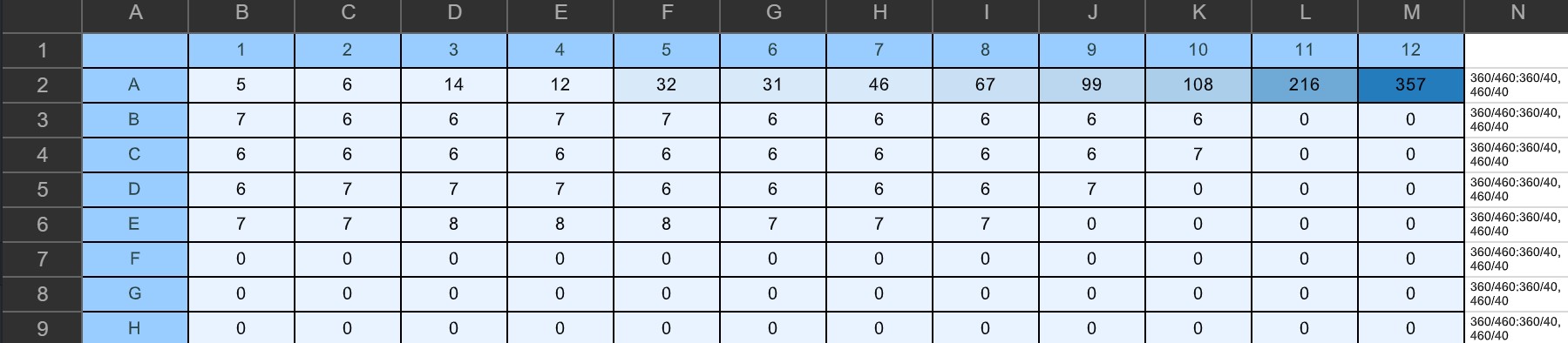
Plate Read Result:
Layout Same as Plate Setup
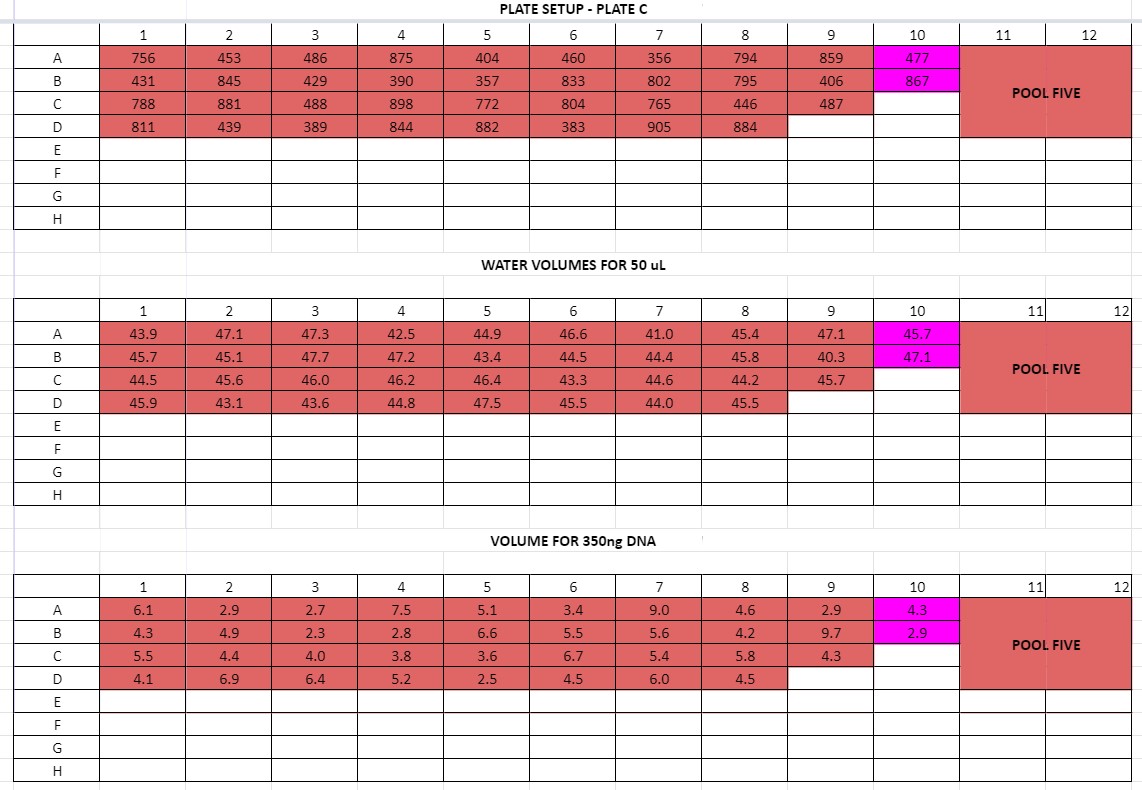
Plate Reader said no DNA present in Plate C, ran a few samples on Qubit to confirm
Samples 887, 760, 422, 769, 403 from plate B for comparison
| Sample | Run 1 |
|---|---|
| S1 | 179.22 |
| S2 | 23037.59 |
| 756 | OUT OF RANGE TOO LOW |
| 845 | OUT OF RANGE TOO LOW |
| 404 | OUT OF RANGE TOO LOW |
| 488 | OUT OF RANGE TOO LOW |
| 833 | OUT OF RANGE TOO LOW |
| 877 | 5.64 |
| 760 | 4.40 |
| 422 | OUT OF RANGE TOO LOW |
| 769 | OUT OF RANGE TOO LOW |
| 403 | 3.86 |
Written on April 27, 2021