Ligation Efficiency Test, Adapter Ligation, Pool 2 Bead Clean
Ligation Efficiency test for Plate B Row A, Adapter Ligation for Plate A, Pooling Pool 2 from Plate B
Date Performed: December 22nd, 2021
Ligation Test for Plate B Row A
Only 1 sample
10ul Hotstart Ready Mix * 2.1 = 21ul
4ul nuclease free water * 2.1 = 8.4ul
.5ul 501 primer * 2.1 = 1.05ul
.5ul 701 primer * 2.1 = 1.05ul
Note:
-
Didn’t move sample to new tube, just broke off technical replicate from rest of strip
- Pipette the ligation test sample(s) into new PCR strip tubes (40ul)
- Perform a 1.5X bead cleanup for each test sample. Briefly:
- Add 60ul room temp KAPA Pure Beads to each sample and pipette mix 10X
- Place on orbital shaker 15 min
- Place on magnet rack and wait for solution to go clear
- Remove clear solution without disturbing beads
- Add 200ul fresh 80% ethanol to tubes
- Remove solution from tubes
- Add 200ul fresh 80% ethanol to tubes
- Remove solution from tubes
- Remove extra liquid with a p20 pipette
- Wait ~2 minutes for excess ethanol to evaporate
- Resuspend beads in 20ul Tris HCL
- Place on orbital shaker for 5 minutes
- Place on magnet rack and wait for solution to go clear
- Remove clear solution from tubes into new strip tubes
- For each ligation test tube, there will be 2 PCR reactions, a 12 cycle and a 30 cycle PCR
- Make a PCR master mix on ice. Thaw ready mix and primers on ice. Vortex and spin down the ready mix and primers before use. Remember each ligation test gets 2 PCRs, so for 1 test samples the “n” number is 2.1 (error), for 2 test samples the “n” number is 4.2, etc. Use any primer set, 50X and 70X pair
- 10ul KAPA HiFi HotStart Ready Mix * “n”
- 4 ul nuclease free water * “n”
- 0.5ul 50X primer * “n”
- 0.5ul 70X primer * “n”
- Make one set of strip tubes for the 12 cycle PCR. Make a second set of strip tubes for 30 cycle PCR
- Into each tube in both strip tube sets that is going to be used (depends on how many test samples you have) add 15ul of the PCR master mix
- Add 5ul from the cleaned test sample(s) into both a tube in the 12 cycle set and the 30 cycle set
- Vortex and spin down the tubes meant for the PCR
- Use two thermocyclers, set the 12 cycle tubes in the thermocycler programed for the 12 cycle RAD LIG TEST program (in JONP Login). Set the 30 cycle tubes in the thermocycler programed for the 30 cycle RAD LIG TEST program
- While that is running, make a small 1% gel to set (gel protocol)
- After the program is done, run 10ul from each PCR reaction (use 2ul dye for 10ul sample) on the gel with a 1kb Plus DNA ladder for 1 hr at 80V You should see a smear on the gel for all of the wells, the 30 cycle samples will be brighter, and might have brighter/more amplification at the smaller fragments (because smaller fragments amplify better in PCRs, which is why we only do 12 cycles in the real prep). The smear shows that the fragments got the adapters so they could be effectively primed and amplified to a level that you can see on the gel. If there is no smear for samples on the gel, it means the ligation did not work and you should go back to step 9 to do the ligation again.

Plate A Adapter Ligation
Ligation master mix for 49 samples\
- 4ul buffer * 49 = 196ul
-
1ul T4 ligase * 49 = 49ul
- Added 5ul LMM to each well
- Added 2ul of correct i5 adapter to each well
- Added 2ul of correct i7 adapter to each well
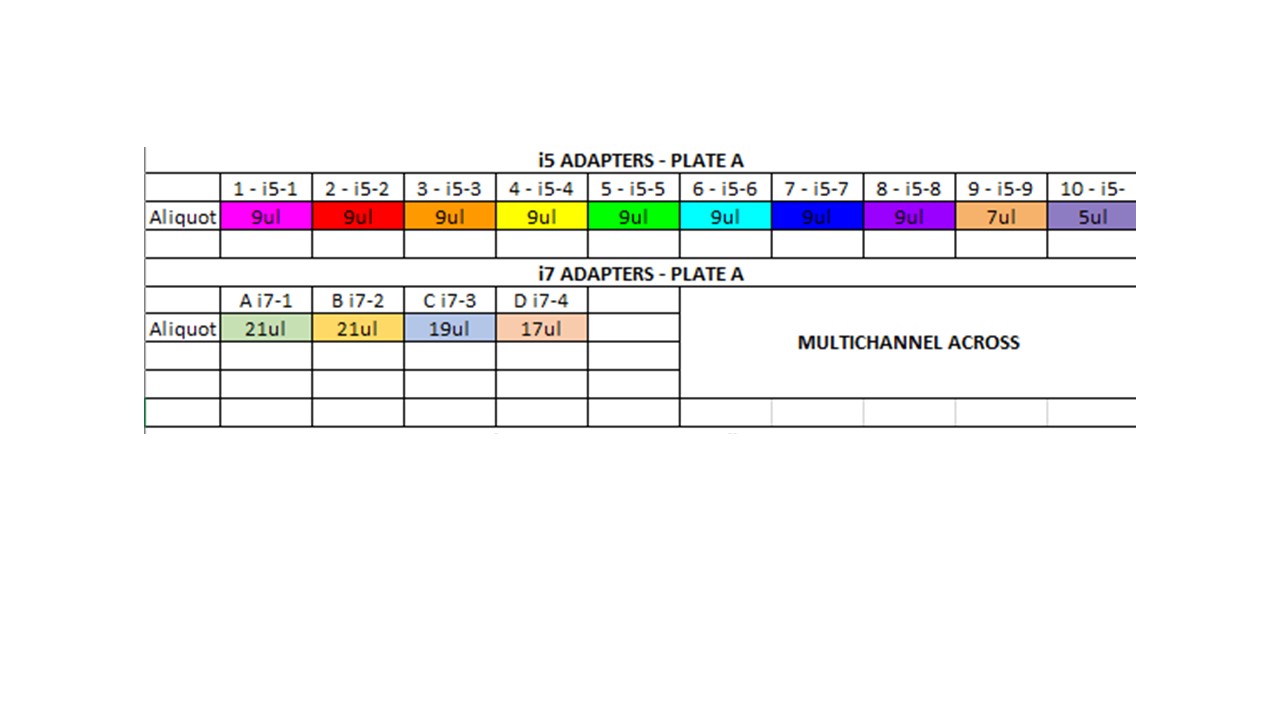
Plate B Pool 2 Pooling and 2 1.5X Bead Cleans
- Take BEADS out of the fridge for ~30 minutes to get to room temperature and make new 80% Ethanol
- For each pool: Transfer the full 40μl of ligation reaction (times the number of wells you are pooling), including the beads to a single 2mL tube. It might help to pipette mix first to make sure to transfer them all. At this stage, do not combine samples from different pools together. It is best to do the pooling and clean ups 1 pool at a time to avoid confusion For each pool: split the 2mL tube into 4 1.5mL tubes with equal volume in each, using a p200 pipette 50μl repeatedly to make sure each tube has the same volume. You can calculate an estimate of the volume in each of the 4 tubes, but usually there has been some evaporation in some wells, so each is not exactly 40ul when you combine them together (you’ll notice this in the step above)
- Add 1.5 times the volume of the 4 tubes of BEADS to each aliquot tube and pipette to mix. This number will change depending on the number of samples per pool
- Incubate the 4 tubes on shaker for 15 minutes
- Place tubes on the magnet bar (long one) and wait until the solution goes clear. You can pipette to encourage the beads to move to the magnet
- Remove as much clear liquid as you can without removing any beads (10-20ul less than the volume in there)
- Add 1000μl 80% EtOH to each tube while keeping it on the magnet
- Remove 1000μl of the EtOH from the tubes and discard
- Add 1000μl 80% EtOH to each tube
- Remove ALL EtOH carefully from each tube, use the p20 or p200 to remove any excess ethanol
- Let samples sit for ~5 minutes, to let the ethanol evaporate but not letting the beads completely dry out
- Remove tubes from magnet and add 40μl of 10mM Tris HCl to each tube, mix by pipetting, and incubate for 5 minutes on the shaker to make sure all the beads are separated from the tube wall
- Stop! Qubit these tubes to make sure you did not lose DNA. Use Qubit Protocol
- You should have about 100ng * how many samples per pool total DNA. To calculate total DNA, multiply the qubit value x 4 x 40.
- Once confirming no dramatic DNA loss, recombine the 4 separated tubes back together into one tube (1 for each pool), this can be one of the tubes you already have samples in
- Add 1.5X BEADS (240μl) to each tube, mix by pipetting, and incubate on a shaker for 5 minutes
- Put the tubes on the magnet, wait until the liquid becomes clear, and remove as much of the clear liquid as you can
- Add 1000μl 80% EtOH to each tube while keeping it on the magnet
- Remove 1000μl of the EtOH from the tubes and discard
- Add 1000μl 80% EtOH to each tube
- Remove ALL EtOH carefully from each tube, use the p20 or p200 to remove any excess ethanol
- Let samples sit for ~5 minutes, to let the ethanol evaporate but not letting the beads completely dry out
- Remove tubes from magnet and add 60μl of 1X TE buffer to each tube and pipette to mix. Incubate for 5 minutes on the shaker
- Put tubes back onto the magnet and wait until the liquid goes clear, then remove supernatant and transfer to a new strip tube and keep on ice
- Repeat these steps for all pools that have gone through the ligation test and are ready to be combined. It is best to cleanup each pool 1 at a time to avoid confusion
- Qubit the pools after the two clean ups to make sure that there is DNA going into the size selection.
- These tubes can now be frozen.
When divided, each of the 4 1.5ul tubes had ~325ul (1.5X clean = 487.5ul beads) Qubit Results for First 1.5X Clean
| Sample | Run 1 | Run 2 |
|---|---|---|
| S1 | 405.85 | |
| S2 | 111549.92 | |
| 1 | 10.2 | 10.2 |
| 2 | 9.92 | 9.88 |
| 3 | 9.88 | 9.88 |
| 4 | 9.74 | 9.72 |
Qubit results from Second 1.5X Clean
| Sample | Run 1 | Run 2 |
|---|---|---|
| S1 | 405.95 | |
| S2 | 110207.97 | |
| Total Pool | 17.4 | 17.2 |
Written on December 22, 2021