Bead Testing
Bead Testing on Old and New Beads
Date Performed: May 6th, 2021
Samples selected for testing: 448-2, 438, 779, 877
Samples from next round of sequencing
- Thawed, vortexed, spun down
- Made two sets of tubes following chart below
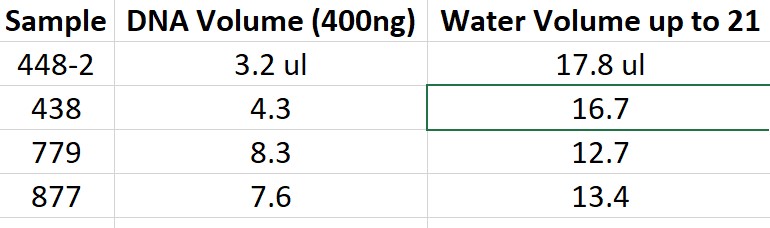
- one is for testing the new beads the other is for testing old beads\
Qubit before clean (after dilution above)
- -N denotes “new” beads, -O denotes “old” beads
| Sample | Run 1 | Run 2 |
|---|---|---|
| S1 | 196.05 | |
| S2 | 22448.42 | |
| 448-N | 26.4 | 26.4 |
| 438-N | 15.9 | 15.4 |
| 779-N | 15.3 | 15.2 |
| 877-N | 29.2 | 28.0 |
| 448-O | 26.6 | 26.2 |
| 438-O | 14.2 | 14.2 |
| 779-O | 14.2 | 14.1 |
| 877-O | 26.0 | 26.8 |
Followed 1.8X bead clean, only change is cleaned with 150ul ethanol instead of 200ul
Eluted in 20ul water because I removed 1ul per sample for Qubit
Post Bead Clean Qubit
| Sample | Run 1 | Run 2 |
|---|---|---|
| S1 | 172.46 | |
| S2 | 22546.19 | |
| 448-N | 23.8 | 23.6 |
| 438-N | 11.8 | 11.8 |
| 779-N | 12.7 | 12.6 |
| 877-N | 18.4 | 18.1 |
| 448-O | 20.2 | 20.2 |
| 438-O | 9.10 | 8.98 |
| 779-O | 9.32 | 9.28 |
| 877-O | 14.8 | 14.5 |
These results didn’t look great. Looked like a lot was lost.
Next I tested three samples by diluting, quantifying, bead cleaning (1.8X), then running in thermocycler.
Added 2ul cutsmart buffer before putting in thermocylcer
| Sample | Volume for 400ng DNA | Volume up to 21ul Water |
|---|---|---|
| 413 | 4.0 | 17.0 |
| 440 | 4.0 | 17.0 |
| 768 | 4.0 | 17.0 |
Pre-Thermocylcer Assay
| Sample | Run 1 | Run 2 |
|---|---|---|
| S1 | 202.64 | |
| S2 | 24478.58 | |
| 413-O | 17.8 | 17.2 |
| 440-O | 20.2 | 19.8 |
| 768-O | 13.5 | 10.6 |
| 413-N | 14.9 | 14.8 |
| 440-N | 20.6 | 20.2 |
| 768-N | 11.0 | 16.4 |
All Results Below (Some duplicate info)
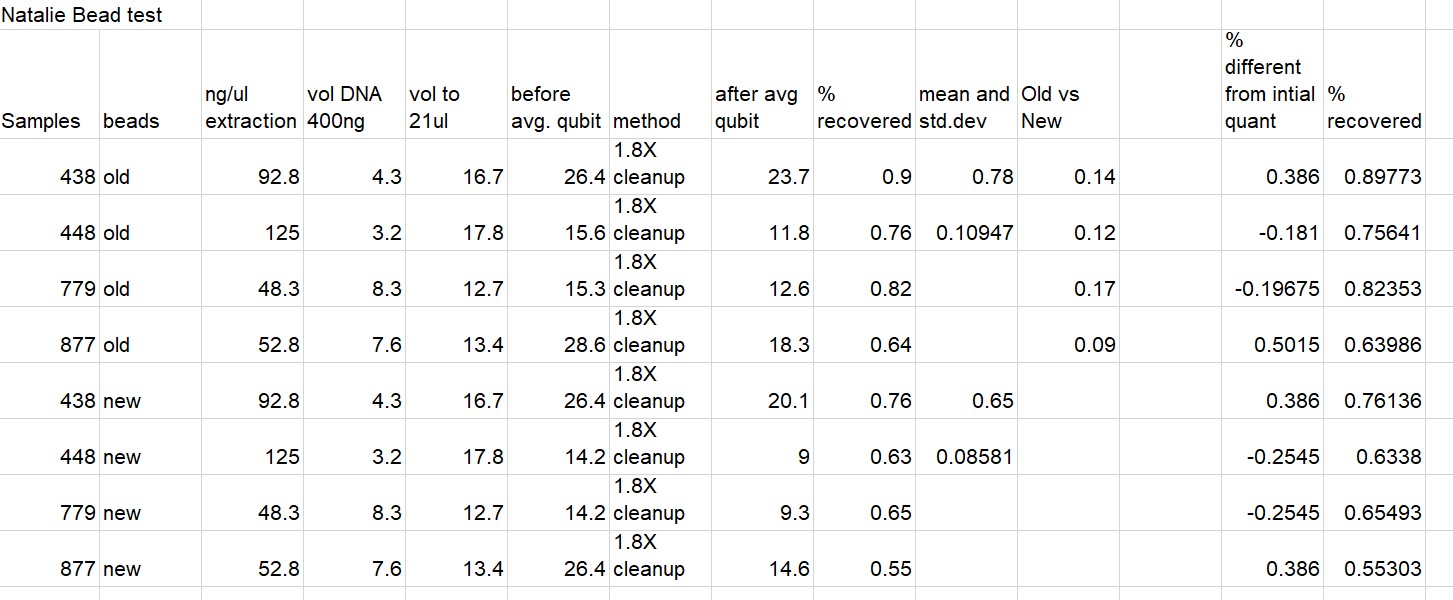
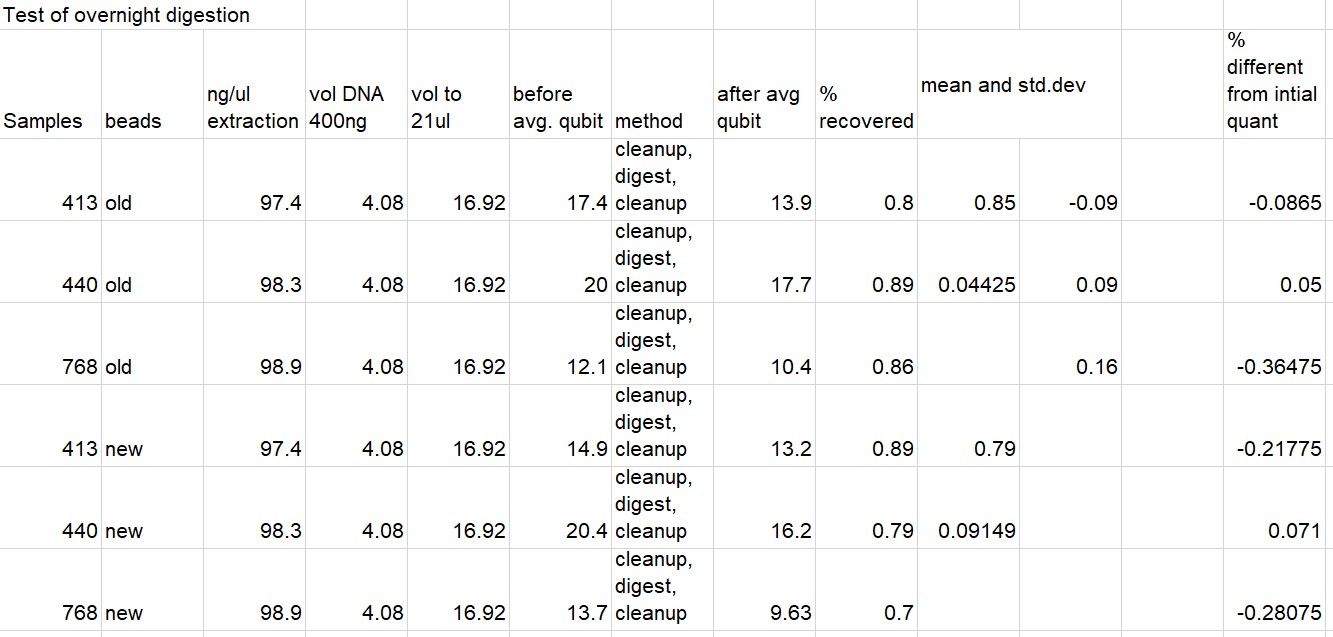
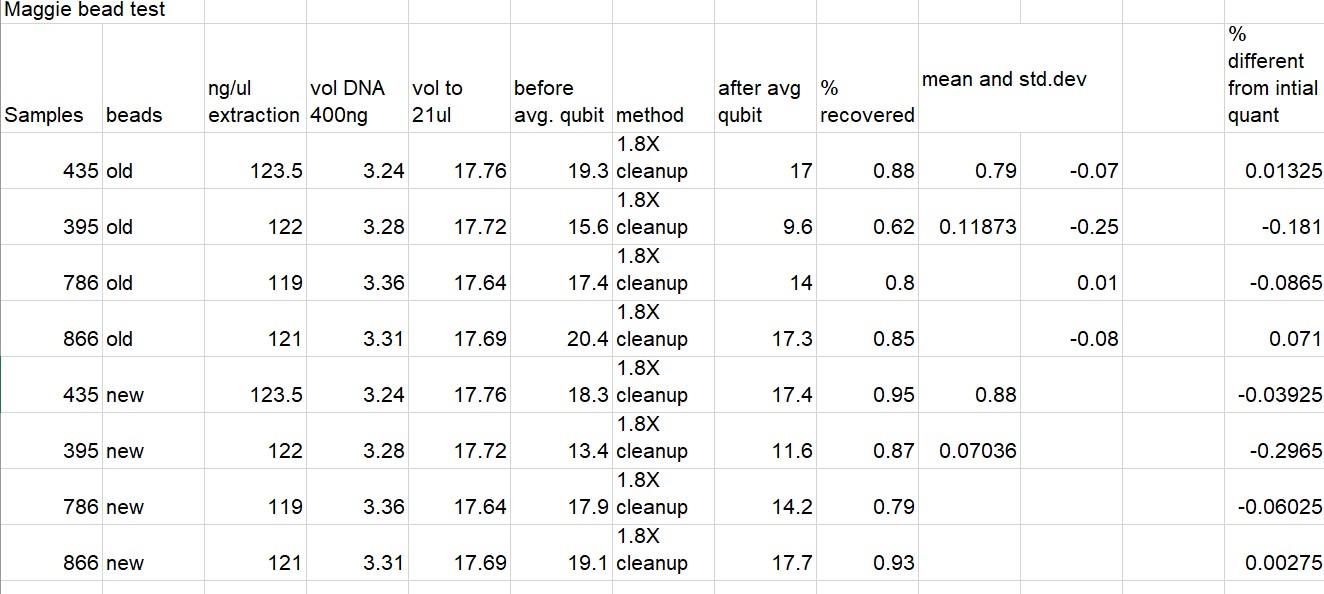
Written on May 6, 2021