Annealing Adapters and Adapter Ligation
Adapter Annealing and Adapter Ligation
Dates Performed: June 29th, 30th, July 1st and 6th
Step 9: Ligation Calculation and adapter annealing and diluting
Plate volume calculations:
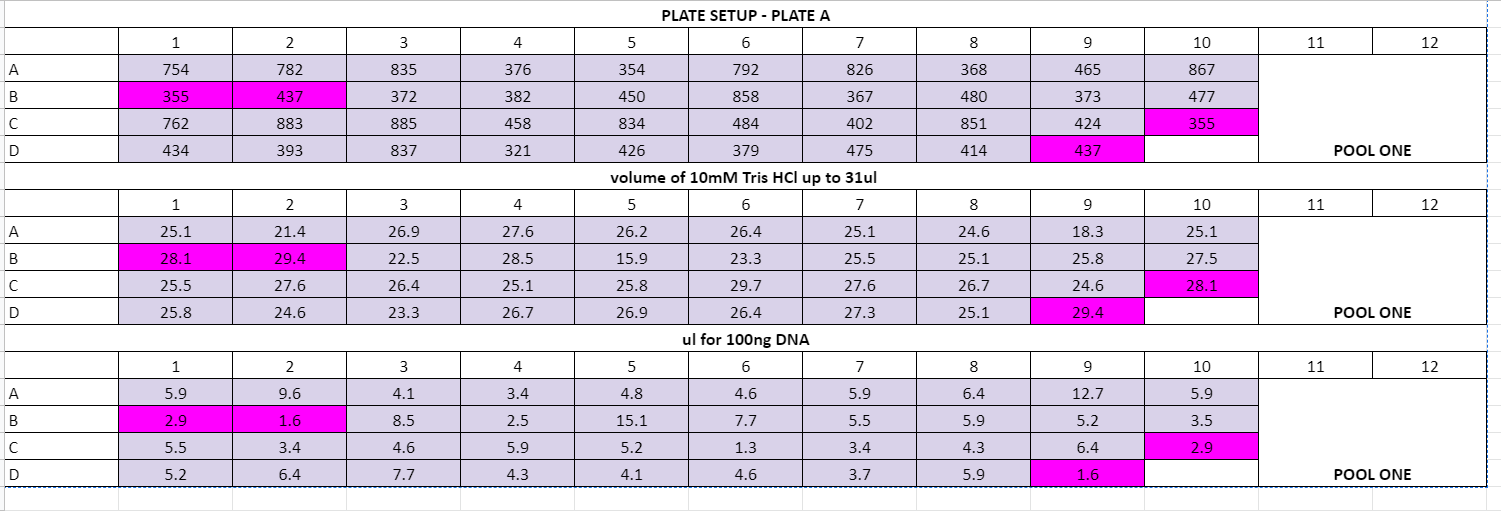
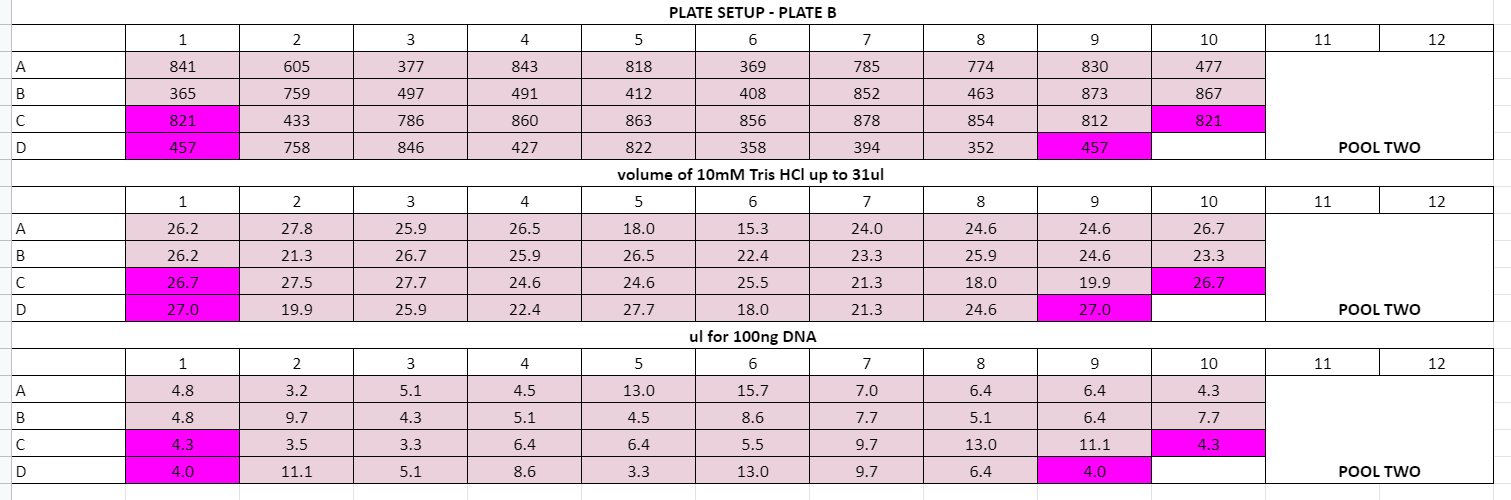
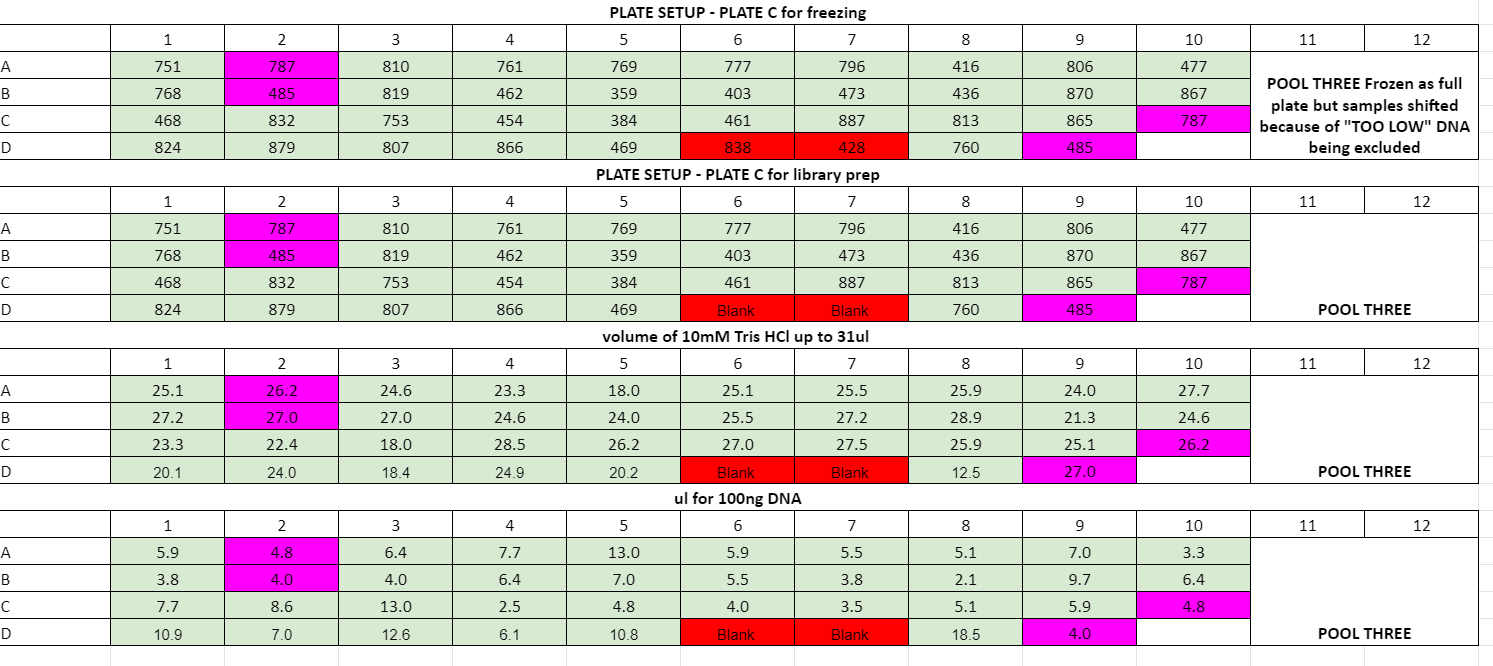
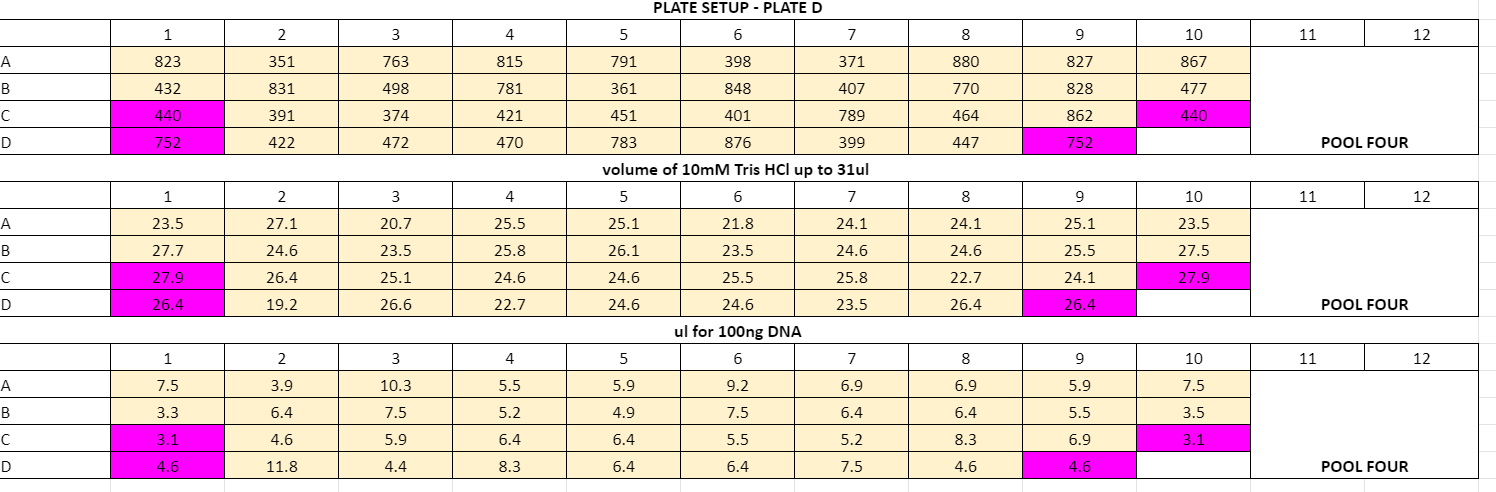
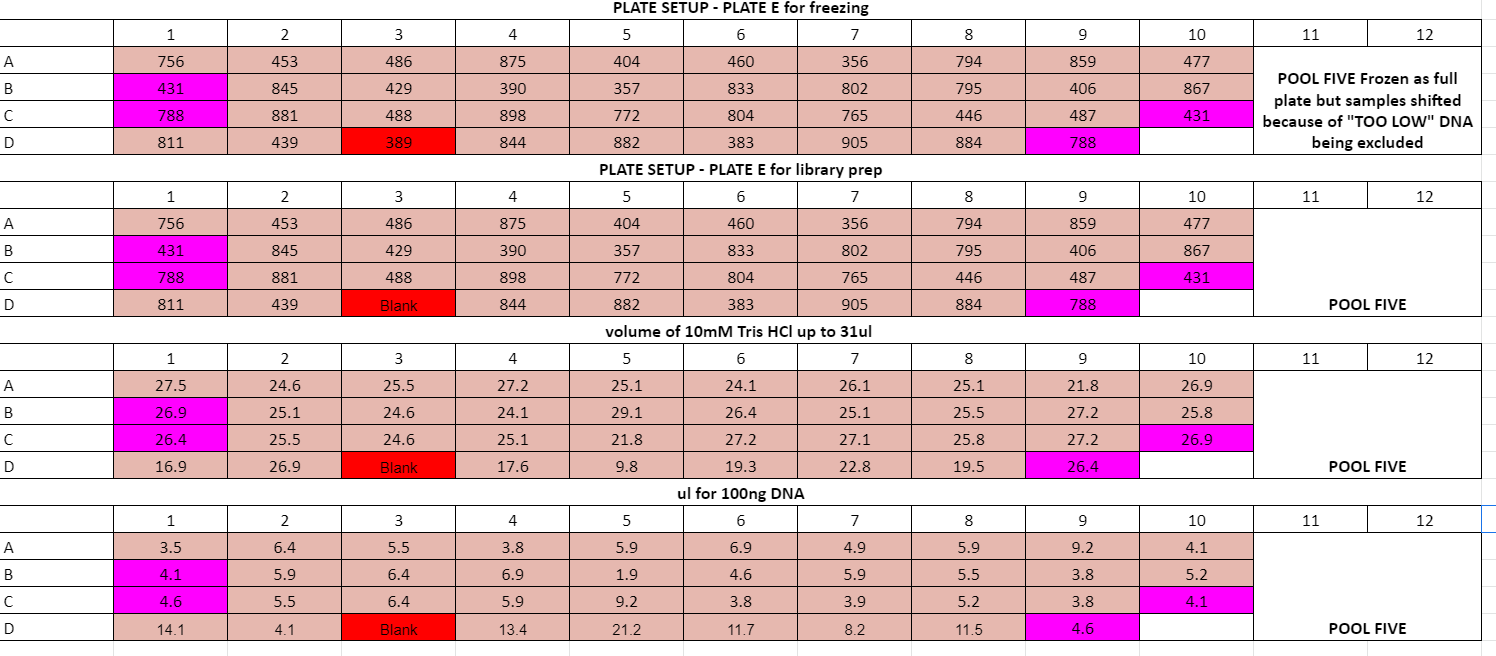
- wells D6 and D7 on plate C had a lot of liquid, I think DNA extraction is good but the DNA got too diluted at some point
- Accidentally added 5ul of i5 adapter to row A of plate D, need to keep in mind for pooling
Anneal adapters and dilute adapters if you have not done so for this project yet Anneal top and bottom adapters into 40uM annealed stock solutions (only had to do this for EcoR1, PST1 already annealed) Make 10X annealing buffer if not made already (in a 50mL conical): 5mL of 1M Tris HCl 5mL 5M NaCl 1mL 0.5M EDTA 39mL Nuclease free water Top and bottom adapters are resuspended to 200uM concentration (usually, check first). Vortex and spin down each before combining For each adapter combine in a strip tube: 10ul top adapter (200uM) 10ul bottom adapter (200uM) 5ul 10X annealing buffer 25ul Nuclease-free water Vortex and spin down and keep on ice Place all adapters to be annealed in the thermocycler adapter annealing program under the JONP login (password 1234) (35 mins)
Dilute adapters to the working stock concentration for your prep. Copy this calculator sheet here and create a new sheet for your project. Clear field B4 and replace with 0.1 (DNA mass for ligation in ug, we are doing 100ng or 0.1ug) Clear fields B6 and B7. Go to the TapeStation results on the TS computer in the analysis software where the single digestions for the enzymes you are using are. For the first enzyme, pick a representative sample single digestion and create a region across the whole distribution. Write down the concentration in ng/ul (in the table below for the region). Convert to pg/ul by multiplying by 1000. That goes in field B6. Write down the molarity as nmol/l (in the table below for the region). Convert to pmol/l by multiplying by 1000. That number goes in B7 Clear fields C6 and C7. Repeat the process in the step above for the second enzyme Look for fields B19, B20, C19, and C20. Those tell you how much of the 40uM annealed stock and 1x annealing buffer to add to a new tube for the working concentration adapters for your project Dilute the 10X annealing buffer by adding 5mL of the 10X annealing buffer to a new 50mL conical and filling it to 50mL with nuclease free water Combine the working stock adapters with the 1X annealing buffer and annealed adapters at the appropriate volume in new strip tubes. Keep these on ice while using and freeze when not in use
PST1 - i5
- anneal adapter stock 3.38ul
- 1X annealing buffer 196.62ul
EcoR1 - i7
- anneal adapter stock 2.08ul
- 1X annealing buffer 197.92ul
Step 10: Adapter Ligation USE FILTER TIPS FOR ALL PIPETTING
Place the plate on the magnet and let the beads go to the magnet Label a new 96 well plate with the plate name and “digested cleaned DNA” Remove the volume of clear liquid in each well to their corresponding well in the new plate (~30ul). Do not pick up the beads Well by well, add back to the bead-plate the volume of DNA for each well for 100ng. The DNA goes back into the same well. Use a plate map to avoid confusion. For the ligation test sample(s), add them in where you planned them to be extra in the plate Add the calculated amount of 10mM Tris HCl needed to increase the volume to 31ul for each well Remove the plate from the magnet and keep on an ice bucket Foil seal the digested DNA plate and freeze at -20 in case you need to do the ligation again later Make the Ligation Master Mix (LMM) on ice. Vortex and spin down the buffer before use. Flick to mix the ligase and spin down. Do not vortex the ligase 4μl of 10X ligation buffer * (number of samples(41)1.05) = 164 1μl of T4 ligase * (number of samples(41)1.05) = 41 Flick to mix the LMM and spin down, keep on ice Aliquot the LMM into strip tubes for multichannel pipetting into each well. Each well gets 5ul of LMM. Use rows or columns, whichever is easier to pipette. Keep on ice (10 strip tubes: 20ul,20,20,20,20,20,20,20,20,15) Aliquot out the adapters into strip tubes for easy addition to your samples (by going down the column and across the rows). Make one set of strip tubes for i5 adapters (added down columns) and make one set of strip tubes for i7 adapters (added across rows). Each well will get 2ul of the correct i5 adapter and 2ul of the correct i7 adapter. Keep on ice Add 5ul of LMM to each well in the plate using the multichannel Add 2ul of the correct i5 indexes by multichannel pipetting down columns Add 2ul of the correct i7 indexes by multichannel pipetting across rows Make sure that the ligation test sample(s) get LMM and adapters, however it does not matter which adapters they get Pipette mix the wells with a p200 pipette set to 25ul Foil seal the plate and spin it down in the large centrifuge Put the plate in the Thermocyclers in the RAD LIGA program in the JONP login (password 1234). This takes between 4-5 hours to run. The program goes to a 4 degree hold afterwards The plate can be taken out and put in the fridge until pooling
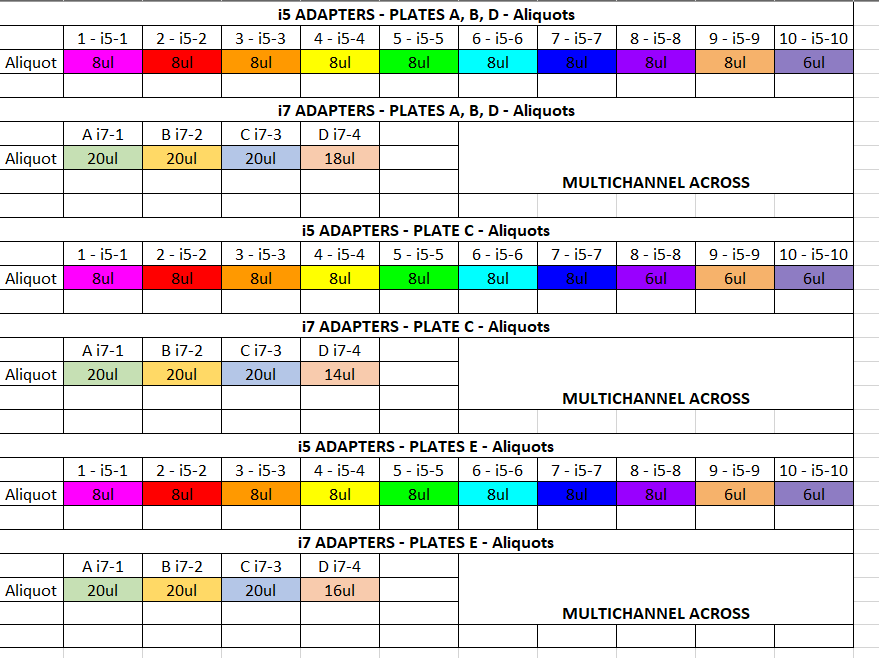
- add 1ul per tube for error
Plates A,B,D processed on 7/1/2021, Plates C and E diluted on 7/1 adapters ligated on 7/6